Science boils down to building models of real-world phenomena 1.
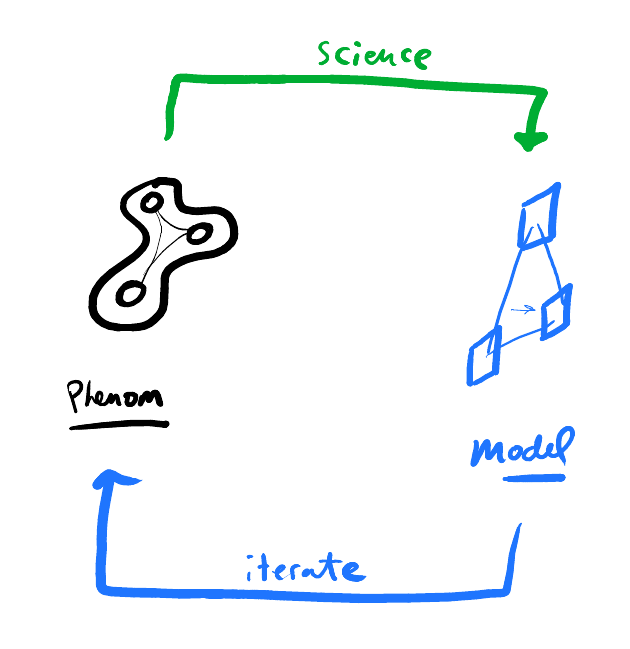
There are, broadly, two ways to do science - piece-by-piece and mode-by-mode.
Piece-by-Piece Science
In Piece-by-Piece (PbP) Science, we break a real-world phenomena, or system, up into its physical components. We then focus on studying interactions between these components.
The typical tool we use here is the experiment - where we try to isolate variables and measure things perfectly.
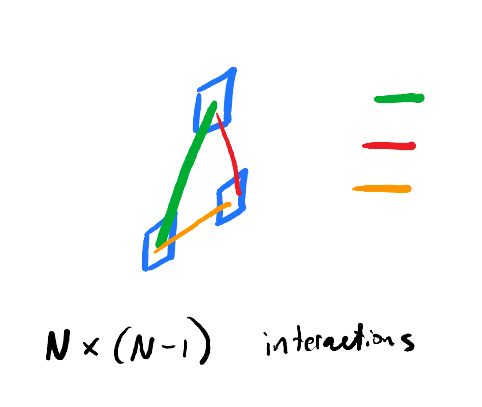
The assumption is that if we study all the interactions between components, we directly have an understanding of the system’s behaviors.
Dyadics
Most efforts in this space focus solely on pairwise, or dyadic, interactions between components. This is what boils down practically to “reductionism”.
More sophisticated efforts here expand to non-dyadic interactions 2. But, across most approaches, we choose a number of components we think are interacting a priori.
Key Point
The key thing, across all of these approaches, is that the PbP approach puts components front stage. The idea is that an understanding of downstream function will naturally emerge as some sort of “sum of interactions”.
Mode-by-Mode Science
In Mode-by-Mode (MbM) Science, we don’t focus on components. We focus on an abstraction one layer above. And then, if we need to, we trace back the ensemble of components involved.
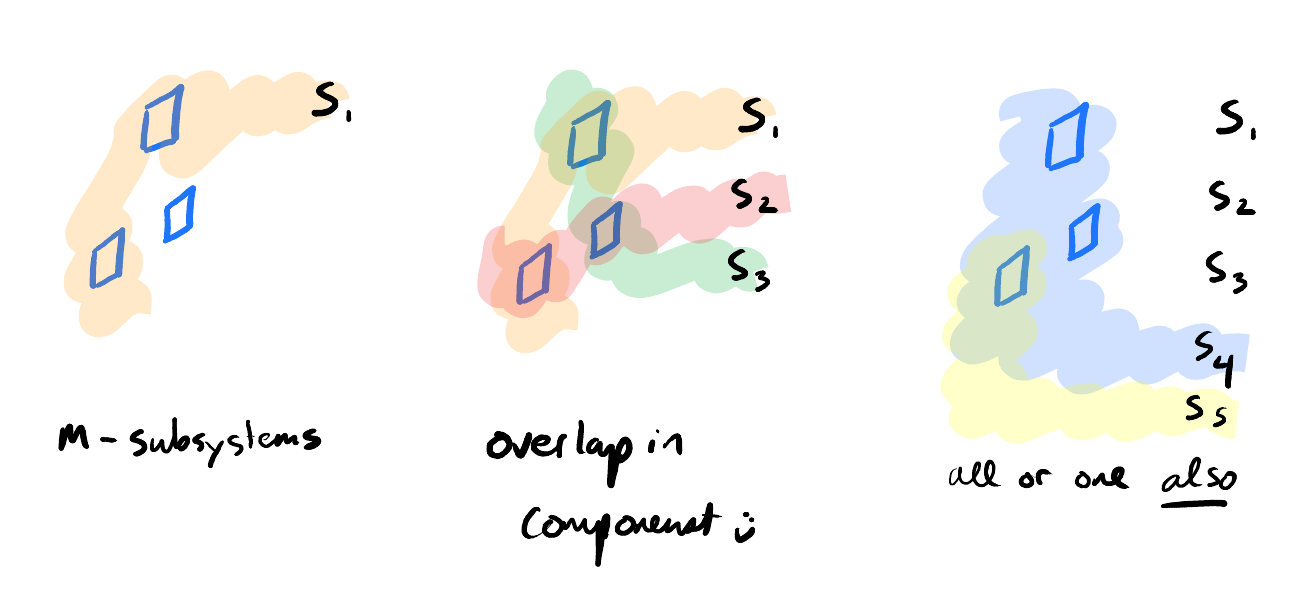
We have flexibility in choosing what we want to focus on - maybe we want to focus on a particular circuit. In which case, we can trace back the components that all mediate a flow of some sort.
Or we want to focus on all components within a particular physical region. In which case, we boil down to a slightly more sophisticated version of our PbP approaches. But one where we have some additional structure that we can leverage to make sense of things more expediently 3.
Reverse Engineering
My favorite type of MbM is reverse-engineering where we focus on a particular subfunction. We find all the parts that dance along with a specific subfunction, then we characterize how all of those parts (a) relate to the function but, more importantly (b) how all of those parts relate to each other.
Much more on this later.
Key Point
The key thing, across all MbM approaches, is that there’s a property, characteristic, or output that takes front stage, and all the parts involved are derived backwards from there.
The central idea is that the added abstraction layer is needed for us to be able to “sum interactions together”, assuming that abstraction layer is structured in an appropriate way.
By setting up a separate layer, we can add in interpretable parameters to impose that structure - giving us the benefit of a sort of “separability” in the spaces we actually care about!
Summary
There are broadly two ways we approach science. Piece-by-Piece focuses on components and how they all interact with each other.
Mode-by-Mode Science focuses on some other abstraction, like subfunction, and then studies the components pulled-back from those subfunctions.
This approach is better for a lot of practical reasons, and is much less likely to miss non-dyadic interaction and even dynamics.
For this post, we’ll take it prima facie. But for more details on this see (my other post)[https://virati.github.io/blog/posts/what_science/]. ↩︎
There’s some great recent work in this respect, but this still remains at the cutting-edge of how we do science. Arguably, the main way contemporary AI can do science is also limited to dyadic interactions. ↩︎
Consider us doing the physical-proximity abstraction - in which case we might find ourselves ignoring long-range interactions since they’re less likely to be strong than short-range interactions. In which case, we could consider it a quicker version of PbP Science. ↩︎